
 Challenges posed by biologial problems are driving the development of new analytical tools and prompting advances in the physical and chemical sciences to help understand the molecular basis of life. Our research covers a wide range of topics, from fundamental questions about the structure and function of biomolecules to understanding the molecular mechanisms of disease. Ph.D. students choose from graduate classes in Biochemistry, Molecular Cell Biology, Molecular and Cell Biophysics, Bio Imaging and Spectroscopy, Molecular Dynamics and Biomolecular Simulation, Biophysics, Molecular Quantum Chemistry, Statistical Thermodynamics, Molecular Spectroscopy.
Challenges posed by biologial problems are driving the development of new analytical tools and prompting advances in the physical and chemical sciences to help understand the molecular basis of life. Our research covers a wide range of topics, from fundamental questions about the structure and function of biomolecules to understanding the molecular mechanisms of disease. Ph.D. students choose from graduate classes in Biochemistry, Molecular Cell Biology, Molecular and Cell Biophysics, Bio Imaging and Spectroscopy, Molecular Dynamics and Biomolecular Simulation, Biophysics, Molecular Quantum Chemistry, Statistical Thermodynamics, Molecular Spectroscopy.
 Many faculty members work at the dynamic interface between chemistry and biology.
Many faculty members work at the dynamic interface between chemistry and biology.
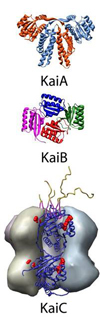
Andy LiWang's group is resolving the structural and biochemical basis of rhythmicity of the circadian clock, which is used for time keeping in life cycles.
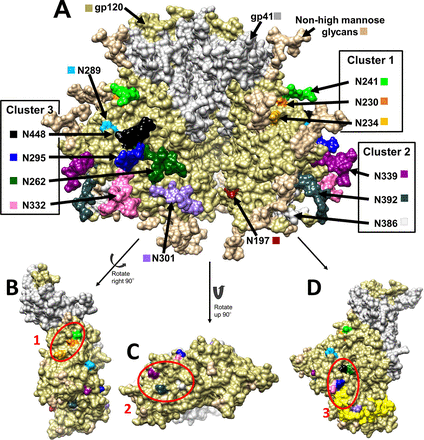
Patti LiWang's group determines the structure of chemokines and how they affect HIV and inflammatory diseases.
Eva de Alba's lab studies the regulation of inflammation and cell death at the molecular level using a variety of biophysical techniques (e.g., NMR, TEM, optical tweezers/fluorescence microscopy) and designs proteins to function as biologics and for different biotechnological applications.
The Noy group studies molecular transport and signal transduction across nanoscale interfaces and develops biomimetic nanostructures that facilitate the creation of bioelectronic devices and circuits.
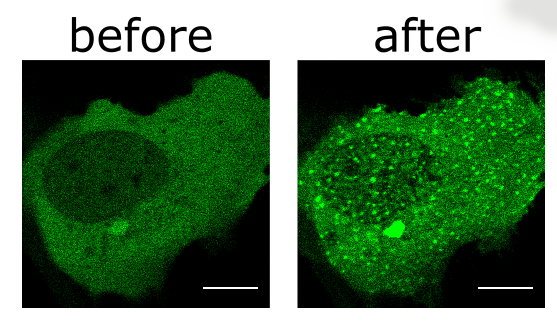 The Ye group is engaged in the hierarchical self-assembly of nucleic acid nanostructures as well as the development of new tools to analyze single DNA molecules.
The Ye group is engaged in the hierarchical self-assembly of nucleic acid nanostructures as well as the development of new tools to analyze single DNA molecules.
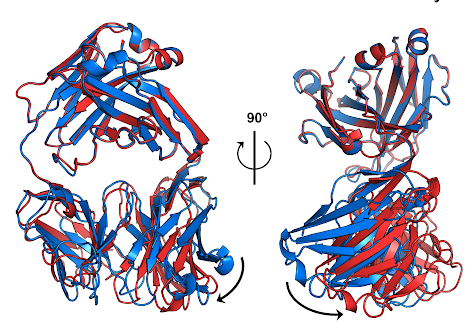 The Thompson Lab is creating new experimental methods that combine temperature perturbations with X-ray crystallography and other techniques, allowing them to explore the conformational landscapes of protein molecules and identify the structural states that define their biological activities.
The Thompson Lab is creating new experimental methods that combine temperature perturbations with X-ray crystallography and other techniques, allowing them to explore the conformational landscapes of protein molecules and identify the structural states that define their biological activities.
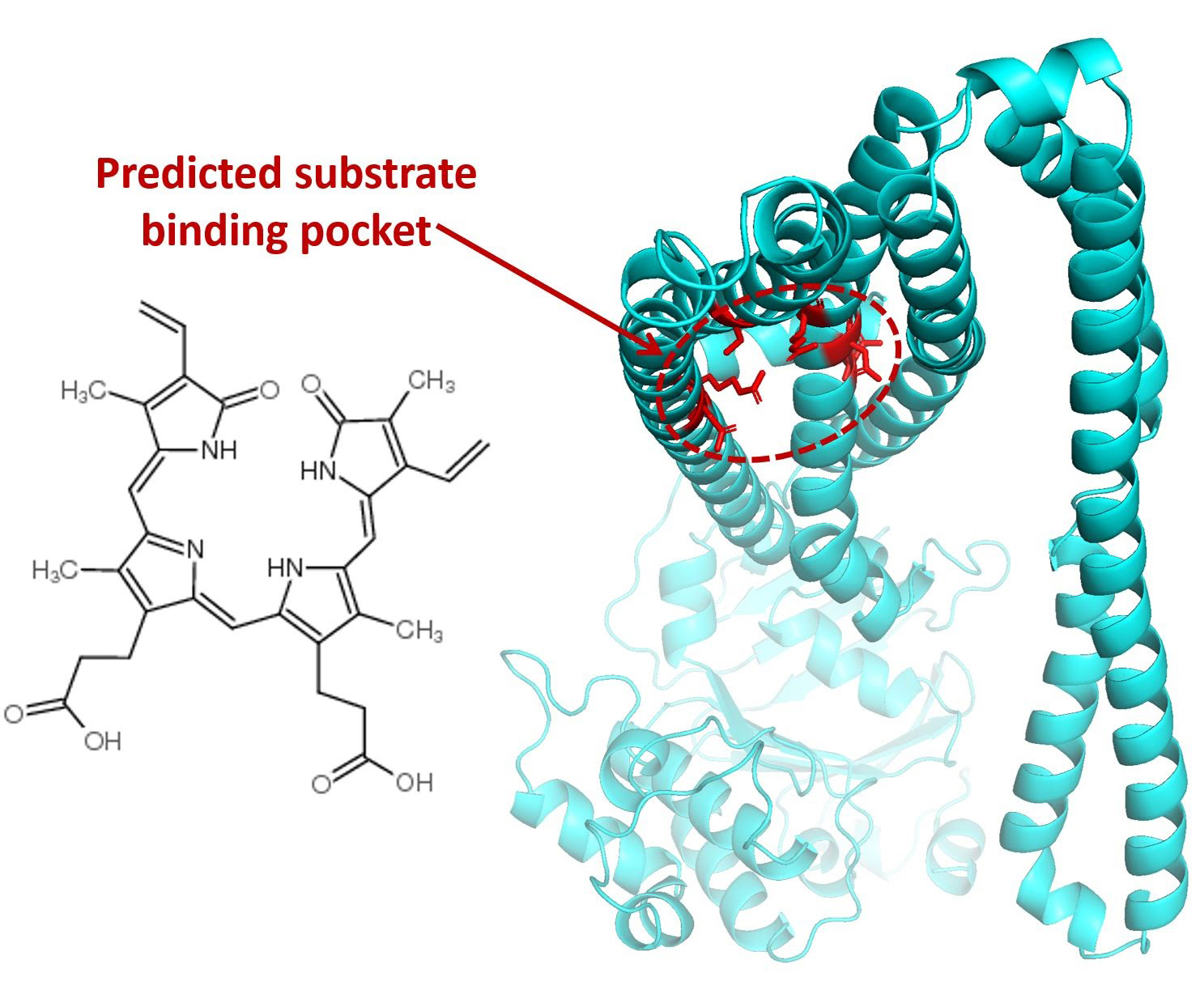 The Zoghbi group studies the structure and function of membrane transport proteins using biochemical, spectroscopic, and electron microscopy techniques.
The Zoghbi group studies the structure and function of membrane transport proteins using biochemical, spectroscopic, and electron microscopy techniques.
The Subramaniam lab is developing new experimental techniques using in vitro synthetic cells to understand fundamental physicochemical mechanisms that govern the assembly and function of biomembranes and proteins.
 Andrea Merg's group is developing synthetic methods for constructing programmable, hierarchical nanomaterials from the self-assembly of designed, molecularly engineered biopolymer building blocks.
Andrea Merg's group is developing synthetic methods for constructing programmable, hierarchical nanomaterials from the self-assembly of designed, molecularly engineered biopolymer building blocks.
The Muñoz group uses experimental and computational protein biochemistry and engineering, and aims to understand protein folding, function and disease and the molecular mechanisms of gene expression, as well as develop novel technologies such as fluorescence biosensors, molecular diagnostics, and allosteric nanoassemblies.



